![]()
Figure 1: Basic principle of SATIC method
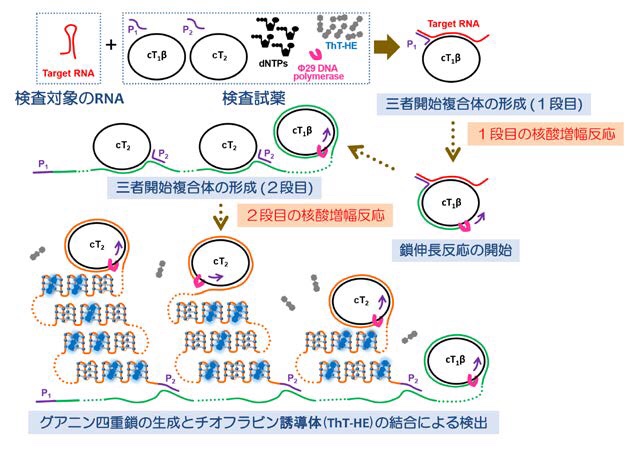
Figure 2: Flow to judgment (procedure)
Figure 3: Test results by PCR method
Figure 4: Test results by SATIC method

COVID-19:塩野義製薬、高感度/ウイルス迅速診断法:唾液から25分で検出:
A Rapid Diagnostic Method with High Sensitivity without a Detection Device in 25 Minutes:
COVID-19: Shionogi,高灵敏度/快速病毒检测方法:在25分钟内检测唾液
塩野義製薬/日本大/群馬大/東京医科大
新型コロナウイルスを含む感染症領域のウイルス迅速診断法で、 日本大学、群馬大学、東京医科大学と業務提携します。
- 唾液等のサンプルから、
- 25分の反応で
- 検出機器を必要とせず
- 目視で判定可能な、
高い感度をもつ迅速診断法です。
塩野義製薬は、日本大学、群馬大学、東京医科大学との間におきまして、新型コロナウイルス(SARS-CoV-2)を含むウイルスの新規迅速診断法に関するライセンス契約に合意しました。
今後、当社は公的機関やアカデミア、パートナー企業と連携し、本診断法の実用化に向けて取り組んでまいります。
従来の検査法:
COVID-19患者を診断する検査法としては、鼻腔や咽頭、唾液から採取した検体からウイルスの核酸を検出するPCR法(ポリメラーゼ連鎖反応)や抗原検査キット等が用いられています。
しかし、これらの検査法では、専用測定器の必要性や、測定の簡便性、迅速性、検体採取時の医療従事者の感染リスク等、依然として多くの課題が残っています。
今回の検査法:
日本大学、群馬大学、東京医科大学からなる共同研究チームが、全く新しい革新的核酸増幅法(SATIC法)によるウイルス迅速診断法の開発に成功しました[1]。
SATIC法は、特定の遺伝子のみならず、変異遺伝子、さらにはタンパク質や代謝物などの生体内分子も、簡便な手法で特異的かつ高感度に測定できる技術です。
本SATIC法を感染症の原因となるウイルスに適用した迅速診断法は、以下の特徴の全てを兼ね備えています。
核酸増幅法(SATIC法):
- ウイルスの感染の有無を、検出機器を必要とせず目視で容易に判定
- 検体採取から25分程度で判定
- 偽陽性反応等の非特異反応がなく、PCR法と同等の高い感度
- 鼻咽頭ぬぐいの綿棒のみでなく、唾液や喀痰からの検出
- 患者の侵襲性が低く、検体採取に伴う感染危険性が限りなく低減
ニュース|シオノギ製薬(塩野義製薬)
https://www.shionogi.com/jp/ja/news/2020/06/200622.html
Business Partnership with Nihon University, Gunma University, and Tokyo Medical University for a Rapid Diagnostic Methods for Viruses in the Field of Infectious Diseases, Including Novel Coronavirus
A Rapid Diagnostic Method with High Sensitivity that can be Visually Determined without the Need for a Detection Device in 25 Minutes of Reaction from Samples such as Saliva
Osaka, Japan, June 22, 2020 –
Shionogi & Co., Ltd.
announced today that Shionogi has agreed with
- Nihon University,
- Gunma University, and
- Tokyo Medical University on a license
agreement regarding a new rapid diagnostic method for viruses including the novel coronavirus (SARS-CoV-2).
We will work in collaboration with public institutions, academia including the above three universities, and partner companies to put this diagnostic method into practical use.
With continued social disruption caused by the worldwide spread of SARS-CoV-2, the spread of infection from not only patients with symptoms but also non-progressor and the infected person in the incubation period has become a major issue in the control of infectious disease.
At present, as the test methods for diagnosing patients with novel coronavirus infection (COVID-19),
PCR (polymerase chain reaction)
are used for diagnosing COVID-19 for detecting nucleic acids in viruses from samples collected from the nasal cavity, pharynx and saliva as well as antigen test kits.
However, many challenges remain with these testing methods, such as
- the need for specific detector,
- the ease and speed of measurement, and
- the risk of infection of healthcare workers
- during sample collection.
Therefore, there is a need for highly sensitive and inexpensive diagnostic methods that overcome these challenges.
The Joint Research Team,
has succeeded in developing a unprecedented innovative viral rapid diagnostic method using signal amplification by ternary initiation complexes method (SATIC method)[1].
The SATIC method is a technology
that can measure not only specific genes, but also mutant genes, as well as in-vivo molecules such as proteins and metabolites, using a simple method with high specificity.
This rapid diagnostic method using SATIC methods has all of the following features.
- The presence or absence of SARS-CoV-2 or influenza virus can be easily determined visually without a detector.
- Virus determination can be made in about 25 minutes after sample collection.
- There is no non-specific reaction such as false positive reaction, and high sensitivity equivalent to that of PCR.
- The ability to detect viruses from saliva and sputum as well as nasopharyngeal swabs reduces the invasiveness of the patient
- the risk of infection among healthcare workers associated with specimen collection as much as possible.
In the case of saliva, specimens can be collected by the patient themselves.
|News|Shionogi Co., Ltd.