Kyoto University: Identifies genes related to Alzheimer’s disease with iPS:
Kyoto University
Professor Haruhisa Inoue (Neurology)
Using “iPS cells made from patients with Alzheimer’s disease”
“Multiple genes involved in Alzheimer’s disease” were identified.
The research team announced by the 21st.
Targeting these genes will lead to early diagnosis and development of therapeutic drugs.
Alzheimer’s disease:
Alzheimer’s disease
The cause is “excessive accumulation of specific proteins in the brain”.
This time, we investigated “sporadic”, which accounts for more than 90% of illnesses and has no family history.
Genetic analysis of blood cells:
The research team
IPS cells made from the blood cells of 102 patients were grown into nerve cells in the brain, and the disease was reproduced at the cellular level.
Identify 24 genes:
Analyze the function of genes in cells.
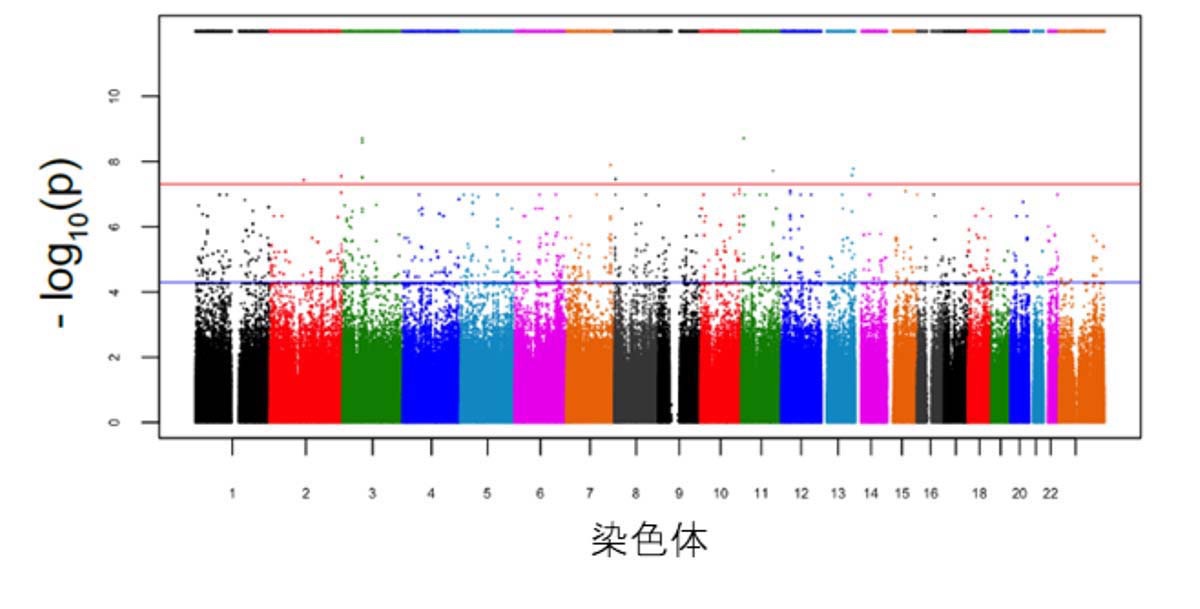
We identified “24 genes that appear to be related to specific proteins”.
Moreover,
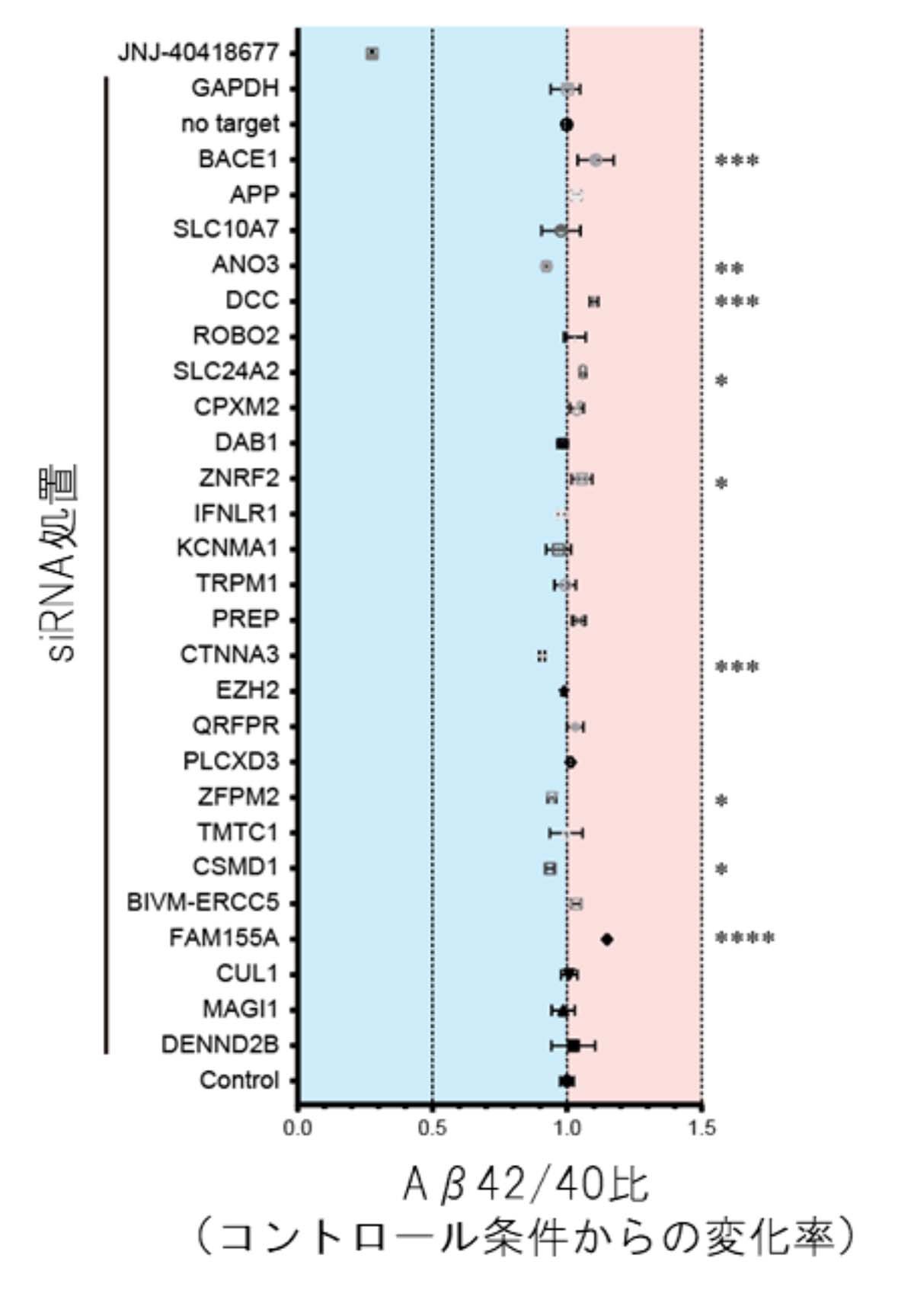
It was also found that “8 of these genes are involved in the regulation of the amount of specific proteins.”
High-precision prediction model:
Utilize information on identified genes and clinical data.
Build a model that predicts the onset of disease with high accuracy from individual genetic information.
Nihon Keizai Shimbun
https://www.nikkei.com/article/DGXZQOUE219O80R20C22A2000000/
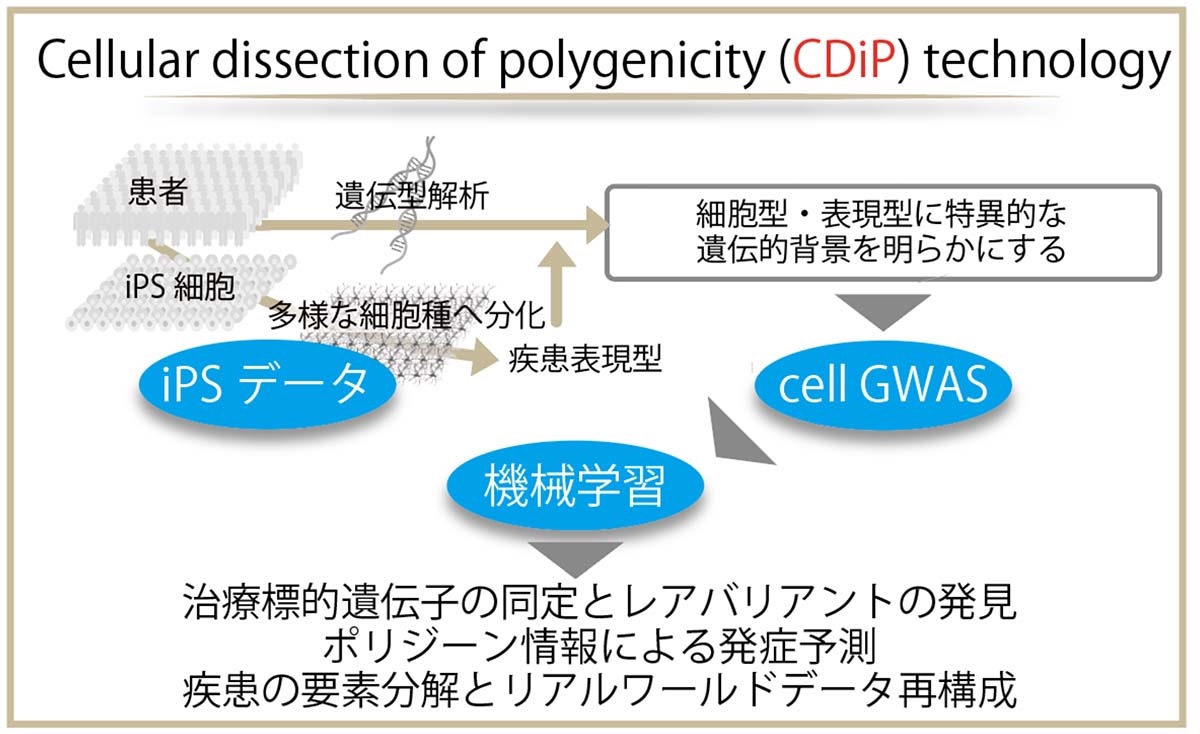
Alzheimer’s disease reconstruction using iPS cohort and machine learning
-Decoding sporadic aging diseases for a disease-free society with CDiP technology-
CiRA | Center for iPS Cell Research and Application, Kyoto University
https://www.cira.kyoto-u.ac.jp/j/pressrelease/news/220218-010000.html
Dissection of the polygenic architecture of neuronal Aβ production using a large sample of individual iPSC lines derived from Alzheimer’s disease patients
Nature Aging
Abstract
Genome-wide association studies
have demonstrated that polygenic risks shape Alzheimer’s disease (AD).To elucidate the polygenic architecture of AD phenotypes at a cellular level,
we established induced pluripotent stem cells from 102 patients with AD, differentiated them into cortical neurons and conducted a genome-wide analysis of the neuronal production of amyloid β (Aβ).
Using such a cellular dissection of polygenicity (CDiP) approach,
we identified 24 significant genome-wide loci associated with alterations in Aβ production,
including some loci not previously associated with AD, and confirmed the influence of some of the corresponding genes on Aβ levels by the use of small interfering RNA.
CDiP genotype
sets improved the predictions of amyloid positivity in the brains and cerebrospinal fluid of patients in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort.Secondary analyses of exome sequencing data from the Japanese ADNI and the ADNI cohorts
focused on the 24 CDiP-derived loci associated with alterations in Aβ led to the identification of rare AD variants in KCNMA1.