
From Japan: Plastic-eating enzyme: Ideonella sakaiensis
Expected to be applied to recycling:
American and British research teams:
The company announced that it has succeeded in “creating an enzyme that can decompose plastics up to 6 times faster than before.”
Discovered by Kyoto Institute of Technology:
Japanese researchers such as Kyoto Institute of Technology have discovered bacteria that break down in a few days after eating PET.
The basis of this research is the discovery reported by a Japanese research team in 2016.
It was discovered at a PET bottle processing factory in Sakai City, Osaka Prefecture.
Researchers have named the bacterium “Ideonella sakaiensis.”
PET = polyethylene terephthalate
PET = polyethylene terephthalate, which is a plastic widely used as a material for PET bottles, has been thought not to decompose in nature.
US-UK research team:
This time, the US and UK research teams
Two enzymes that break down PET found in Sakaiensis in two steps are artificially bound.
I made a “super enzyme”.
“Using this, it was confirmed that PET can be decomposed up to 6 times faster than before.”
TV Tokyo NEWS
https://news.yahoo.co.jp/articles/4d5794f45c4eb6f44da83a9f35b15a3259de8342
(PNAS) Brandon C. Knott et al.(2020)Characterization and engineering of a two-enzyme system for plastics depolymerization
Abstract
Plastics pollution
represents a global environmental crisis. In response, microbes are evolving the capacity to utilize synthetic polymers as carbon and energy sources.
Recently, Ideonella sakaiensis
was reported to secrete a two-enzyme system to deconstruct polyethylene terephthalate (PET) to its constituent monomers.
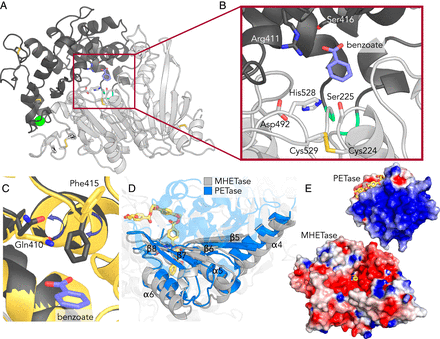
Specifically, the I. sakaiensis PETase depolymerizes PET, liberating soluble products, including mono(2-hydroxyethyl) terephthalate (MHET), which is cleaved to terephthalic acid and ethylene glycol by MHETase.
Here, we report a 1.6 Å resolution MHETase structure, illustrating that the MHETase core domain is similar to PETase, capped by a lid domain.
Simulations of the catalytic itinerary predict that MHETase follows the canonical two-step serine hydrolase mechanism.
https://www.pnas.org/content/early/2020/09/23/2006753117
EurekAlert! Plastic-eating enzyme ‘cocktail’ heralds new hope for plastic waste
https://www.eurekalert.org/pub_releases/2020-09/uop-pe092520.php